Autores
Camargo-ayala, P.A. (UNIVERSIDAD DE TALCA) ; Cofré-leiva, C. (UNIVERSIDAD DE TALCA) ; Vergara, A. (UNIVERSIDAD DE TALCA) ; Hernández-olivos, R. (UNIVERSIDAD DE TALCA) ; Zúñiga-hernández, J. (UNIVERSIDAD DE TALCA) ; Sanhueza, S. (UNIVERSIDAD DE CONCEPCIÓN) ; Nova-lamperti, E. (UNIVERSIDAD DE CONCEPCIÓN) ; Rivera, C. (UNIVERSIDAD DE TALCA)
Resumo
There are currently no alternatives to prevent recurrent aphthous stomatitis
(RAS). The objective of this research was to characterize the salivary proteome of
patients with RAS, we used saliva from patients both in the active phase and in
recurrence comparing it with healthy patients, to be analyzed through proteomics
based on mass spectrometry. We found that the ATF6B protein was able to
differentiate between patients in the active phase and remission, several proteins
related to cell death, were also concentrating centers DPP4, a target receptor for
several drugs. Our data suggest late apoptotic cell death and the pathological
role of chemokines. We suggest a possible therapeutic target for future research.
Palavras chaves
proteomics; aphthous stomatitis; mass spectrometry-based p
Introdução
Recurrent aphthous stomatitis (RAS) is the most common ulcerative disease of the
oral mucosa. It is characterized by very painful multiple or single recurrent
lesions (Chavan.2012). Unlike other common oral diseases, it cannot be
prevented. The lesions usually resolve between one and two weeks after their
appearance and in part this has contributed to underreporting. Current evidence
strongly supports that the immune system is activated in RAS. The lesions are
preceded by a T helper cell hyperimmune response (Brocklehurst.2012). To date,
the reason that leads to its activation and, ultimately, to the development of
RAS is unknown. Saliva could be used as an approximation for the measurement of
disease-related biomarkers (Nonaka.2022). RAS is a condition that goes through
phases of activity and inactivity. There are even people who never have injuries
(Esteves.2019). Exploring what distinguishes the subjects who suffer from it can
guide the nature and biological bases of this disease (Rivera.2019). Taking
these factors into account, the aim of this study was to examine the salivary
proteome of RAS patients using proteomics and computational biology. To do this,
we conducted a crossover case study, analyzing salivary samples from healthy
controls and patients with RAS throughout the ulcerative cycle (active ulcers
and absence of lesions). Our findings revealed a response that tries to avoid
the destruction of oral keratinocytes, as well as the possibility that the types
of cell death involved in the disease are apoptosis or paraptosis. Furthermore,
data mining, gene ontologies, protein-protein interactions, and network
approaches suggest a potential therapeutic target.
Material e métodos
Our research is a cross-case study. We evaluated the salivary proteome of RAS
patients during the presence and absence of ulcers together with healthy
controls using a mass spectrometer and bioinformatics tools. This study was
authorized by the Ethics Committee of the University of Antofagasta #156/2018.
After obtaining informed consent, we collected 118 salivary samples from 68
subjects. Participants were divided into two groups: healthy controls (n=31;
people with no history of RAS ulcers) and people with RAS (n=36). This last
group was evaluated at the beginning of the study, during the ulcerative stage
and when the lesions completely disappeared (remission stage, n=36). A nanoElute
LC system paired with a timsTOF Pro-BD mass spectrometer was used, mass
spectrometry data was obtained by examining 500 ng of peptides. All MS/MS
samples were analyzed with PEAKS Studio X+ according to the above protocols
(Fraga.2021). We describe the main biological and molecular processes of each
group using the FunRich software enrichment analysis (Pathan.2015). Spectral
count values were used to determine differential protein abundance between
conditions in Perseus software (Lundgren.2010-Tyanova.2016). Western and dot-
blot were performed to identify ATF6B (Yang.2020-Barrera.2016). In addition, we
measured TNF-α and IFN-γ cytokines using the Cytokine Bead Array Th1/2/17 BD kit
and flow cytometry. To identify genes that encode proteins related to apoptosis,
we used the Génie web tool (Fontaine.2011). Prioritized genes were analyzed
using CellPhoneDB (Efremova 2020-Szklarrczyk 2016-Kearney-2018). we enumerate
the selected receptors in the database we call the resulting network "RAS cell
death interactome".
Resultado e discussão
The salivary proteome of patients with RAS was studied. The ATF6B protein made
it possible to distinguish between samples with lesions and those in which the
oral mucosa had not been destroyed (Rivera.2020). We also identified an anti-
cell death response and a potential therapeutic target, DPP4. In RAS there is
evident death of oral keratinocytes and it seems that the type of death points
to apoptosis mainly due to the detection by mass spectrometry of salivary ATF6B.
Proteomic results were verified by western blot and dot blot assays, where ATF6B
was more expressed in the presence of RAS ulcers. Excessive ER stress activates
several apoptotic pathways (Chadwick.2019).The presence of salivary ATF6B in
subjects with ulcerative lesions may be a marker of ER stress response in oral
keratinocytes. ATF6B expression may indicate that ER stress must be reduced for
re-epithelialization (progress to remission stage) (Bachar.2021), to occur. In
terms of cell signaling, the major group of protein-protein interactions shows
that CCL2 and CCL5 with more connections (Armutc.2013). The switch that
initiates the RAS mechanisms is still unknown. Revealing the molecules that
constitute the main centers of activity in the RAS apoptosis interactome may
provide an opportunity to find new therapies. Here, DPP4 emerged as a
participant in the RAS cell death interactome with three drugs that can inhibit
its action DPP4 can influence lymphocyte function in a variety of ways
(Kleman.2016), including T-cell activation and signal transduction DPP4 is
thought to have functions immunoregulatory (Shao.2020), as well as a therapeutic
promise in the treatment of autoimmune and inflammatory diseases. Together with
our results, they make DPP4 an interesting molecule to evaluate in future
experiments.
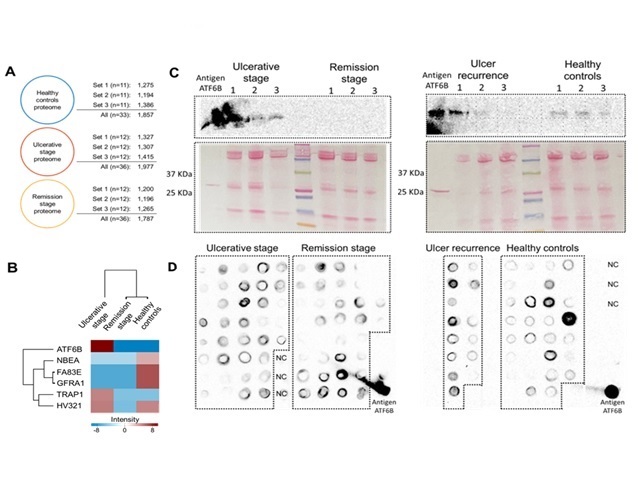
(A) Number of saliva proteins (B)ATF6B stands out (C)Western blot (D)Dot blot
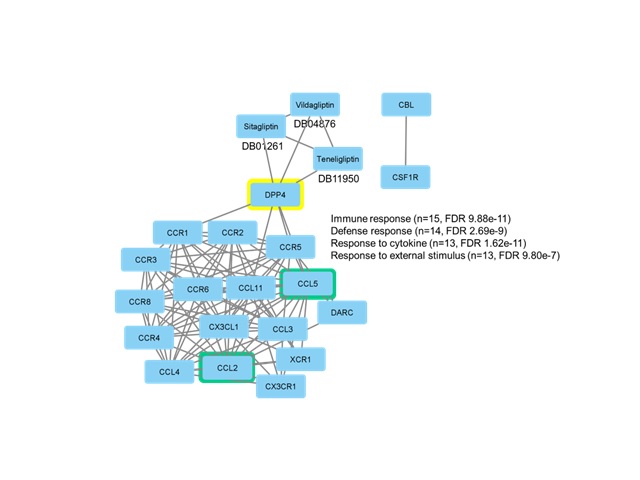
Network with proteins processed by STITCH and its pharmacological relationship with drugs for the treatment of Type 2 diabetes mellitus.
Conclusões
Salivary proteomics allowed to identify proteins that allow to distinguish between
groups of subjects with and without ulcers. The ATF6B protein suggests that the
trigger for keratinocyte death affects the normal functioning of the ER, causing a
late apoptosis-type death. Cell death process using computational biology
identified a network of highly connected proteins, one is a therapeutic target for
FDA-approved drugs. From a clinical perspective, this could open new options for a
more specific treatment for RAS.
Agradecimentos
We thank the Melissa Institute. Financing was provided by ANID, Scholarships:
21220448-P.A.C-A, 22221111-C.C-L and FONDECYT: 1211480-E.N.-L., 11200258-J.Z-H and
11180170-C.R.
Referências
BARRERA, MJ. Pro-inflammatory cytokines enhance ERAD and ATF6α pathway activity in salivary glands of Sjögren’s syndrome patients. J. Autoimmun. 75, 68–81 (2016).
BROCKLEHURST, P. et al. Systemic interventions for recurrent aphthous stomatitis (mouth ulcers). Cochrane Database Syst. Rev. CD005411 (2012) doi:10.1002/14651858.CD005411.pub2.
CHAVAN, M. et al. Recurrent aphthous stomatitis: a review. J. Oral Pathol. Med. 41, 577–583 (2012).
EFREMOVA, M., VENTO-TORMO, M., TEICHMANN, S. A. & VENTO-TORMO, R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506 (2020).
ESTEVES, C. V. et al. Diagnostic potential of saliva proteome analysis: a review and guide to clinical practice. Braz. Oral Res. 33, e043 (2019).
FONTAINE, J.-F., PRILLER, F., BARBOSA-SILVA, A. & ANDRADE-NAVARRO, M. A. Génie: literature-based gene prioritization at multi genomic scale. Nucleic Acids Res. 39, W455–61 (2011).
FRAGA, M. et al. Immunomodulation of T Helper Cells by Tumor Microenvironment in Oral Cancer Is Associated With CCR8 Expression and Rapid Membrane Vitamin D Signaling Pathway. Front. Immunol. 12, 643298 (2021).
KEARNEY, P., BONIFACE, J. J., PRICE, N. D. & HOOD, L. The building blocks of successful translation of proteomics to the clinic. Curr. Opin. Biotechnol. 51, 123–129 (2018).
LUNDGREN, D. H., HWANG, S.-I., WU, L. & HAN, D. K. Role of spectral counting in quantitative proteomics. Expert Rev. Proteomics 7, 39–53 (2010).
NONAKA, T. & WONG, D. T. W. Saliva Diagnostics. Annu. Rev. Anal. Chem. 15, 107–121 (2022).
PATHAN, M. et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 15, 2597–2601 (2015).
RIVERA, C. Essentials of recurrent aphthous stomatitis. Biomed Rep 11, 47–50 (2019).
SZKLARCZYK, D. et al. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 44, D380–4 (2016).
TYANOVA, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016).
YAN, G., ELBADAWI, M. & EFFERTH, T. Multiple cell death modalities and their key features (Review). World Acad Sci J (2020) doi:10.3892/wasj.2020.40.
YANG, H., NIEMEIJER, M., VAN DE WATER, B. & BELTMAN, J. B. ATF6 Is a Critical Determinant of CHOP Dynamics during the Unfolded Protein Response. iScience 23, 100860 (2020).
















