Autores
Vélez Vargas, L.C. (INSTITUTO TECNOLÓGICO METROPOLITANO) ; Pedroza Díaz, N.J. (INSTITUTO TECNOLÓGICO METROPOLITANO) ; Henao Castañeda, I.C. (UNIVERSIDAD DE ANTIOQUIA)
Resumo
Wnt pathway dysregulation is associated with many diseases, and the scientific community has been searching for the modulation of its components as a promising therapeutic target. The inhibition of the PORCN and DVL does not allow the transduction of signals, consequently affecting the transcription of proliferation genes. This study aims to determine the binding affinity and interactions between CGA and the targets DVL and PORCN by molecular docking. Here was found that CGA shows an affinity for the mentioned proteins, and the way of interaction is like the validation compounds. Therefore, it is necessary to study structural modifications of the CGA to improve the inhibitory affinity for these proteins.
Palavras chaves
molecular docking; dishevelled; porcupine
Introdução
The Wnt signaling pathway is critical because it regulates cell proliferation, migration, differentiation, survival, and adhesion (AMADO, PREDES, et al., 2014), by controlling β-catenin and consequently the expression of key genes (CHENG, XU, et al., 2019). Due to its role, the dysregulation of the signaling is associated with many diseases as cancer, neurodegenerative disorders, fibrosis, endocrine diseases and metabolic syndrome (SFERRAZZA, CORTI, et al., 2020). For this reason, molecular components of the pathway have been proposed as innovative therapeutic targets.
Wnt's are cysteine-rich glycoproteins secreted as ligands to promote receptor-mediated signal transduction pathways. These proteins undergo a characteristic modification in which an acyl group from palmitoleic acid is attached by the palmitoyl transferase Porcupine (PORCN) (SFERRAZZA, CORTI, et al., 2020). In the absence of Wnt ligands, β-catenin is degraded by a multimeric protein complex, consisting on APC, Axin, GSK3β and CK1, responsible for preventing the accumulation of it in the cytoplasm (CHENG, XU, et al., 2019). However, in the presence of Wnt ligands, these molecules interact with Frizzled which also interacts with Dishevelled (DVL) at the PDZ domain (GRANDY, SHAN, et al., 2009). Consequently, occurs a stabilization of β-catenin by disrupted complex; it accumulates and translocate to the nucleus where it engages the TCF/LEF family to mediate transactivation of target genes (CHENG, XU, et al., 2019).
Despite the existence of DVL and PORCN’s inhibitors, the development non-peptidic small compound modulators is necessary. For this reason, this study aims is the analysis of the affinity and interactions between chlorogenic acid (CGA) and two targets of the Wnt/β-catenin signaling pathway.
Material e métodos
DVL and PORCN structures were obtained from Protein Data Bank, code 7URC and 6ZC8, each one with a reported inhibitor, compound 18 (C18) an LGK974 respectively. Proteins were prepared using UCSF Chimera (PETTERSEN, GODDARD, et al., 2004) for eliminate the molecules that were not of interest. Then with AutoDock Tools (ADT) (MORRIS, HUEY, et al., 2009) were remove water molecules, and added only polar hydrogens, AD4 type of atoms and Gasteiger charges. CGA anion, was built using GaussView 5.0 (NIELSEN, HOLDER, 2009) and optimized with GAUSSIAN 09 (FRISCH, TRUCKS, et al., 2016) and B3LYP/6-31++G (d,p) approximation. Validation and interest compounds where prepared using ADT for assigned charges.
For stablish the center were chosen the amino acids implicated on the binding site, for the grid on DVL’s case were use Phe259, Gly261, Ile262, Ser263, Ile264 and Val316, near these was the atom CBD from C18, which coordinates (x = 40.459, y = -22.667, z = 17.080) were selected. On the other hand, for PORCN’s were use Phe246, Val297, Trp300, Thr328, Tyr329, Leu346, Leu349 and Leu408, and near these was atom C07 from LGK974, which coordinates (x=110.344, y = 107.969, z = 106.954) were selected.
Molecular docking was carried out in Autodock Vina (TROTT, OLSON, 2009). Re-docking test < 2 Å was performed to evaluate the reproducibility of docking calculation (LI, LIANG, et al., 2020). The pose with the best affinity for each protein-ligand complex was evaluated using Discovery Studio Visualizer (BIOVIA, [S.d.]) and PLIP (SALENTIN, SCHREIBER, et al., 2015). Also, a visual inspection and measurement of distances were performed in UCSF Chimera.
Resultado e discussão
We found that RMSD for Porcn was 1.244 Å, demonstrating that parameters such as center and size of the grid were suitable for molecular docking. Nevertheless, the RMSD for DVL was never under 2 Å. Although this criterion was not accomplished, the analyzes were continue, since the interactions that the inhibitor formed with the PDZ domain of DVL coincide with those reported in the literature.
CGA presented less affinity to both proteins than the validation compounds (Tables 1 and 2). In the PORCN case, LGK974 forms more hydrophobic interactions than CGA, associated with aromatic rings, while CGA forms more hydrogen bonds due to hydroxyl groups. Furthermore, C18 is higher in size than CGA, allowing more interactions with DVL binding site.
LIU, QI, et al., mentioned that LGK974 formed hydrophobic interactions with residues described at the establishment of the grid’s center. Also was observed that residue Ser332 interacting with the carbonyl oxygen of LGK974. We confirmed all the interactions described above except the interaction with Leu349. Also, hydrophobic interactions with Leu308, Leu342, and Phe412 were found. On the other hand, interactions with Tyr329, Trp300, and Thr328 turned to hydrogen bonds when CGA is in the binding site, while interactions with Val297 and Leu346 conserved its hydrophobic nature.
KAMDEM, ROSKE, et al., reported that their compounds form hydrogen bonds with residues Leu260, Gly261, Ile262 and C18 form a π-π stacked interaction with Phe259. In this study, we confirmed these interactions between ligands C18 and CGA with PDZ. Additionally, we found hydrophobic interactions with Val316 and Leu319 between C18 and DVL. Meanwhile, between CGA and DVL were found hydrogen bonds with Ile264 and Asn312, and hydrophobic interactions with Val316.
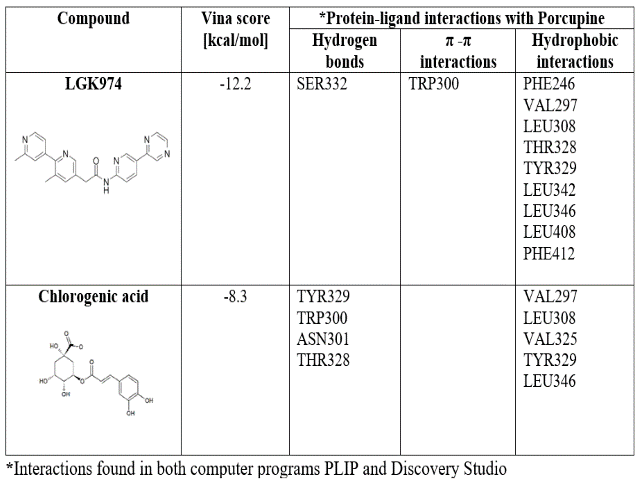
Molecular docking results for biding between Chlorogenic acid and LGK974 with Porcupine.
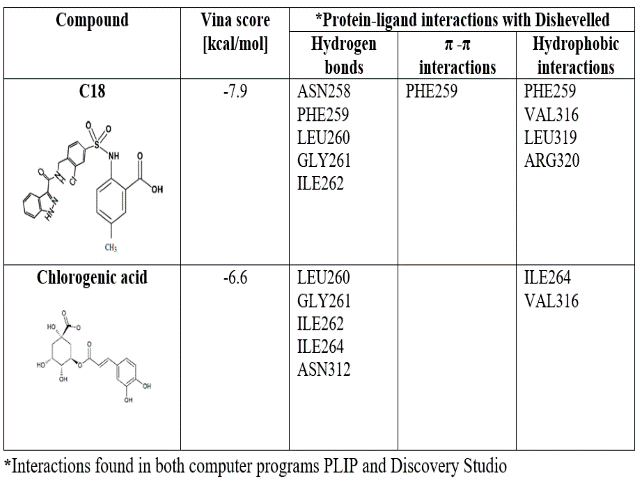
Molecular docking results for biding between Chlorogenic acid and LGK974 with Dishevelled.
Conclusões
Our findings demonstrate that CGA can interact with PORCN and DVL similarly to the validation compounds LGK974 and C18, respectively. Nevertheless, there are differences in the binding affinity, probably due to the hydrophobic interactions. Due to these findings, we consider that structural modification to CGA to achieve higher lipophilicity could improve the affinity for PORCN and DVL binding sites. Additionally, there might be in vitro studies that prove the protein-ligand interaction at the evaluated sites.
Agradecimentos
This work was supported by the Instituto Tecnológico Metropolitano and by a Minciencias grant (project code: 115080763215 CT 811-2018).
Referências
AMADO, N. G., PREDES, D., MORENO, M. M., et al. "Flavonoids and Wnt/β-catenin signaling: Potential role in colorectal cancer therapies", International Journal of Molecular Sciences, v. 15, n. 7, p. 12094–12106, 2014. DOI: 10.3390/ijms150712094. .
BIOVIA. Discovery studio visualizer. . [S.l.], Dassault Systèmes. , [S.d.]
CHENG, X., XU, X., CHEN, D., et al. "Therapeutic potential of targeting the Wnt/β-catenin signaling pathway in colorectal cancer", Biomedicine and Pharmacotherapy, v. 110, p. 473–481, 2019. DOI: 10.1016/j.biopha.2018.11.082. Disponível em: https://doi.org/10.1016/j.biopha.2018.11.082.
FRISCH, M. J., TRUCKS, G. W., SCHLEGEL, H. B., et al. Gaussian 09. . Wallingford, CT, USA, Gaussian Inc. , 2016
GRANDY, D., SHAN, J., ZHANG, X., et al. "Discovery and characterization of a small molecule inhibitor of the PDZ domain of dishevelled", Journal of Biological Chemistry, v. 284, n. 24, p. 16256–16263, 2009. DOI: 10.1074/jbc.M109.009647. .
KAMDEM, N., ROSKE, Y., KOVALSKYY, D., et al. "Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling", Magnetic Resonance, v. 2, n. 1, p. 355–374, 2021. DOI: 10.5194/mr-2-355-2021. .
LI, B., LIANG, J., LU, F., et al. "Discovery of Novel Inhibitor for WNT/β-Catenin Pathway by Tankyrase 1/2 Structure-Based Virtual Screening", Molecules, v. 25, p. 1–17, 2020. .
LIU, Y., QI, X., DONNELLY, L., et al. "Mechanisms and inhibition of Porcupine-mediated Wnt acylation", Nature, v. 607, n. 7920, p. 816–822, 2022. DOI: 10.1038/s41586-022-04952-2. .
MORRIS, G. M., HUEY, R., LINDSTROM, W., et al. "AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility", Computational chemistry, p. 2785–2791, 2009. .
NIELSEN, A. ., HOLDER, A. . GaussView 5. . [S.l: s.n.]. , 2009
PETTERSEN, E., GODDARD, T., HUANG, C. C., et al. "UCSF Chimera--a visualization system for exploratory research and analysis.", Computational chemistry, 2004. .
SALENTIN, S., SCHREIBER, S., HAUPT, V. J., et al. "PLIP: fully automated protein-ligand interaction profiler", Nucleic acid research, 2015. .
SFERRAZZA, G., CORTI, M., BRUSOTTI, G., et al. "Nature-derived compounds modulating Wnt/β-catenin pathway: a preventive and therapeutic opportunity in neoplastic diseases", Acta Pharmaceutica Sinica B, v. 10, n. 10, p. 1814–1834, 2020. DOI: 10.1016/j.apsb.2019.12.019. .
TROTT, O., OLSON, A. J. "Software News and Update AutoDock Vina : Improving the Speed and Accuracy of Docking with a New Scoring Function , Efficient Optimization , and Multithreading", Computational chemistry, n. 31, p. 455–461, 2009. .
















